International bZIP Research Group
From regulatory complexity to biological function:
Metabolic adjustment of plant development by regulatory bZIP factor networks
A Cooperative International Project funded since 2007 by:
Deutsche Forschungsgemeinschaft (DFG)
Netherlands Organisation for Scientific Research (NWO)
Fonds zur Förderung der wissenschaftlichen Forschung Österreich (FWF)
Spanish Ministry of Education and Science (MEC)
Participating Members (PIs) and Contact
Wolfgang Dröge-Laser (alternate coordinator)
Albrecht-von-Haller-Institut
Universität Göttingen
Untere Karspüle 2
D-37073 Göttingen
Phone: +49-(0)551-39-19816
E-mail: wdroege(a)gwdg.de
www.ubpb.gwdg.de/wdllab/index.html
Presentation Utrecht March 2008 (Password)
Klaus Harter (coordinator)
ZMBP / Plant Physiology
University of Tübingen
Auf der Morgenstelle 32
72076 Tübingen
Germany
Phone: +49-(0)7071-29-72605
E-mail: klaus.harter(a)zmbp.uni-tuebingen.de
www.zmbp.uni-tuebingen.de/PlantPhysiology/index.html
Sjet Smeekens
Molecular Plant Physiology
Utrecht University
Padualaan 8
3584 CH Utrecht
The Netherlands
Phone. 030-2533431 (direct) 2534230 (secr.)
Fax. 030-2532837
E-mail: j.c.m.smeekens(a)bio.uu.nl
Markus Teige
Department of Biochemistry
Max F. Perutz Laboratories
University of Vienna
Dr. Bohrgasse 9/5
A-1030 Vienna
Austria
Phone: +43-1-4277-52815 or -52810
E-mail: markus.teige(a)univie.ac.at
www.univie.ac.at/ibmz/groups/teigeov.htm
Jesus Vicente Carbajosa
Dep. Biotecnologia
ETSI Agronomos
Universidad Politecnica de Madrid
Ciudad universitaria s/n
28040 Madrid
Spain
Phone: +34-91-3363261
E-mail: jesus.vicente(a)upm.es
Aim of Our Research
Introduction
A current emphasis in plant molecular biology research is on the control mechanisms that govern metabolism and that integrate metabolism with developmental and responses to the environment. Changes in primary and secondary metabolism often precede or even cause alterations in the growth and development of higher plants in response to endogenous and environmental conditions. Well-known examples are the production and accumulation of protective metabolites combined with reduced growth in plants after exposure to abiotic or biotic stress or the influence of carbohydrates on light-induced seedling photomorphogenesis. In addition, the lack of knowledge of the genetic and molecular basis of the reprogramming of plant metabolism, resource allocation and, thus, yield is hampering traditional and molecular breeding efforts to improve plant productivity.
Although substantial research efforts have been made to understand plant metabolic pathways, we are remarkably lacking in knowledge of the control mechanisms governing plant metabolism and coordinating it with plant development and responses to the environment.
bZIP Transcription Factors
Two groups of basic leucine zipper (bZIP) transcription factors from Arabidopsis form the centre of our research approach (Figure 1). Preliminary genetic and molecular studies by the participating groups have shown that these transcription factors are involved in the fine-tuning of gene expression. Hence, bZIP factors may act in signal response pathways controlling, for instance, the carbon, nitrogen organic acid and reactive oxygen species (ROS) metabolism.
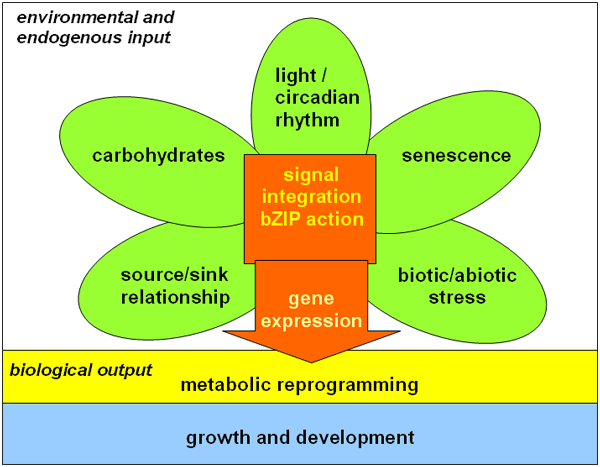
Figure 1. Basic scientific concept of the research project: Basic Leucine Zipper (bZIP) transcription factors act as central integrators of environmental and endogenous input signals in plants. Multiple signals are perceived in parallel and have to be integrated by diverse sets of signalling components (e.g. receptors, kinases) and/or transcriptional regulators. bZIP transcription factors serve as an example here to study metabolic reprogramming and adaptation of growth and development in plants as response to changes in their environment.
Consequently, these bZIP factors could play a crucial role in several developmental processes in higher plants, such as seed maturation, stamen development and senescence and in adaptive responses to the environment, such as to light and abiotic and biotic stresses (Figure 1). Accordingly these groups of transcription factors present an excellent example for studying these complex regulatory processes at different levels, as they provide a wide range of experimental possibilities to address functional consequences in a biological context. Due to the diverse biological processes in which bZIP factors are involved, a comprehensive investigation of these phenomena can only be achieved within a close cooperation of specialists working on different fields of signal transduction and transcriptional regulation.
Selected Publications
Ehlert, A., Weltmeier, F., Wang, X., Smeekens, S., Vicente-Carbajosa, J., Dröge-Laser, W. (2006).
Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerisation map of group-C and S bZIP transcription factors. Plant J. 46, 890-900.
Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S (2008)
The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53, 935-949.
Pubmed
Kaminaka, H., Näke, C., Epple, P., Dittgen, J., Schütze, K., Chaban, C., Holt, B.F., Merkle, T., Schäfer, E., Harter, K.*, Dangl, J.L.* (2006).
Pathogen-induced cell death control via antagonistic functions of a bZIP transcription factor and LSD1 in Arabidopsis thaliana. EMBO 25, 4400-4411.
Schütze K, Harter K, Chaban C (2008)
Post-translational regulation of plant bZIP factors. Trends Plant Sci., Epub ahead of print.
Pubmed
Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., Dangl, J., Hirt, H. (2004).
The MKK2 Pathway Mediates Cold and Salt Stress Signaling in Arabidopsis. Mol. Cell 15, 141-152.
Weltmeier, F., Ehlert, A., Mayer, C.S., Dietrich, K., Wang, Schütze, K., Alonso, R., Harter, K., Vincente-Carbajosa, J., Dröge-Laser, W. (2006).
Combinatorial control of proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 25, 3133-3143.
Benetka W, Mehlmer N, Sammer M, Neumüller R, Maurer-Stroh S, Koranda M, Knoblich J, Teige M, Eisenhaber F (2008).
Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle 23, 3709-3719.
Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schütze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, Vicente-Carbajosa J, Smeekens S, Dröge-Laser W (2009).
Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol. 69, 107-119.
Rahmani F, Hummel M, Schuurmanns J, Wiese-Klinkenberg A, Smeekens S, Hanson J (2009).
Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Phys. 150, 1356-1367.
Hummel M, Rahmani F, Smeekens S, Hanson J (2009).
Sucrose-mediated translational control. Ann. Bot. 104, 1-7.
Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009).
A pivotal role of the basic leucine zipper transcription factor AtbZIP53 in the induction of seed gene expression based on bZIP heterodimerisation and protein complex formation. Plant Cell, doi: 10.1105/tpc.108.062968.
*Shared last authorship
