Biosynthesis of Pristinamycin
The streptogramin antibiotic pristinamycin, produced by Streptomyces pristinaespiralis, is a mixture of two types of chemically unrelated compounds: pristinamycin PI and PII, which are produced in a ratio of 30:70. Pristinamycin PI is a cyclic hexadepsipeptide, belonging to the B-group of streptogramins (Fig. 4), while pristinamycin PII has the structure of a polyunsaturated macrolactone of the A-group of streptogramins (Fig. 4).

Fig.4: Structure of pristinamycin I and II with respective precursor molecules.
Both compounds alone inhibit the protein biosynthesis by binding to the peptidyl transferase domain of the 50S subunit of the ribosome and are bacteriostatic. Together they show a strong synergistic bactericidal activity, which can reach 100 times of the separate components. We showed that the genes responsible for pristinamycin biosynthesis are not organized as a single cluster but spread over 210 kb together with a type II PKS gene cluster (Fig. 5). Currently, we functionally characterize this ’gene region’ by knock-out mutagenesis, protein expression/enzyme assays and other molecular techniques.

Fig. 5: Schematic representation of the pristinamycin gene region. PI biosynthetic genes (red), PII biosynthetic gene (blue), genes for regulation and resistance, and of unknown function (black). The 90 kb interjacent region is shown as a dotted line.
Cryptic pristinaespiralis polyketide
Within the 210 kb pristinamycin gene region we identified a further gene cluster with a size of ~40 kb (Fig. 6). This cryptic pristinaespiralis polyketide (cpp) gene cluster shows high similarity to gene clusters of aromatic polyketides, such as those of granaticin of S. violaceoruber, actinorhodin of S. coelicolor A3(2) and mithramycin of S. argillaceus, which all belong to the class of benzoisochromanequinones. At the moment we try to express this gene cluster homologously in S. pristinaespiralis and heterologously in S. lividans to later on characterize the secondary metabolite encoded by the cpp cluster.

Fig. 6: Schematic representation of the cryptic polyketide gene cluster (cpp).
Phenylglycin biosynthesis
Within the pristinamycin biosynthetic gene region we identified a set of genes (pglA, pglB, pglC, pglD and pglE) putatively involved in L-phenylglycine biosynthesis (Fig. 7A), which is an aproteinogenic amino acid and component of PI (Fig. 7B). By inactivation of the individual genes and feeding experiments with phenylglycine, we could show that the genes are responsible for L-phenylglycine biosynthesis. Currently, we are investigating the phenylglycine biosynthetic pathway by protein expression and enzyme assay experiments.
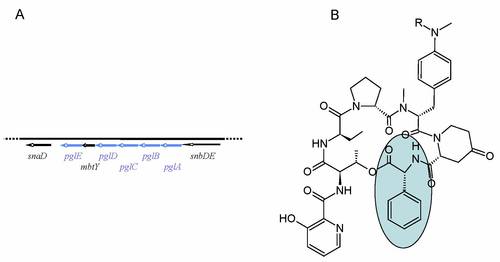
Fig. 7: Schematic representation of the phenylglycine biosynthetic gene operon (A). Structure of pristinamycin I. The aproteinogenic amino acid L-phenylglycine is marked by a blue box (B).
Regulation of Pristinamycin Biosynthesis
In streptomycetes, the regulation of antibiotic biosynthesis is quite complex. The biosynthesis mostly starts with the switch from primary to secondary metabolism. Herby, the presence of certain signal molecules as the γ-butyrolactones (GBL) play an important role. GBL are responsible for the induction of secondary metabolism in streptomycetes and are recognized and bound by the γ-butyrolactone receptor proteins which represents global regulators of antibiotic biosynthesis. In the absence of GBL, the receptor acts as repressor. In the presence of GBL, the repressor looses its ability of DNA binding and transcription of the till then repressed genes can start. Often, further regulatory genes are controlled by the GBL receptor as the so called ‚Streptomyces Antibiotic Regulatory Proteins’ (SARPs). SARPs are pathway-specific regulators which mostly are responsible for the direct activation of all antibiotic biosynthetic genes (abg). Thus, a coordinated expression of the genes is possible. But often, the regulation of antibiotic biosynthesis is more complex. So, several activators as well as repressors may be inserted between GBL receptor and SARPs so that a signal cascade results.

Fig. 8: Simplified presentation of the regulation of antibiotic biosynthesis.
Within the 210 kb pristinamycin biosynthetic gene region, seven regulatory genes were identified: One γ-butyrolactone receptor gene, 3 SARP genes, 2 repressor genes, as well as one gene encoding a response regulator. To investigate the function of those regulatory genes, especially their hierarchy and interaction, deletion mutants and strains overexpressing one or more of the regulators are constructed. Furthermore, we are performing gel shift assays and RT PCR experiments with the aim to elucidate the regulatory signal cascade controlling pristinamycin biosynthesis.
