News
03.03.2021
Protein sequence clustering with DIAMOND
In her bachelor thesis, Jasmin Katz extended and optimized the clustering algorithm of DIAMOND.
There has been an exponential increase in protein sequences in the last decades due to large-scale projects aiming at sequencing unknown species. The number of protein sequences will continue to increase in the future due to large-scale projects such as the Earth BioGenome Project. Protein sequence clustering plays an essential role in analyzing this large amount of data efficiently. This method enables the reduction of large protein datasets and the identification of functional and evolutionary similarities between different proteins.
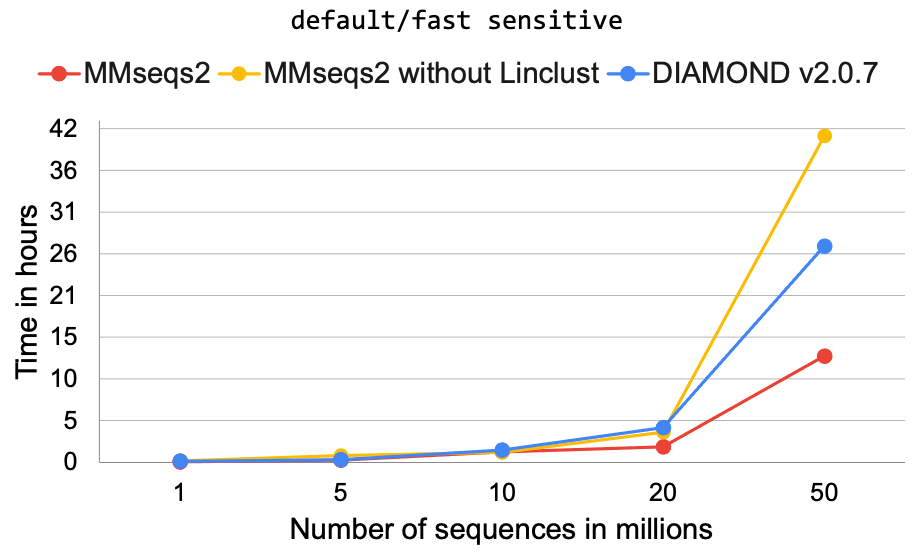
The graph-based Greedy Vertex Cover algorithm used for clustering was extended by the option of cascaded clustering in the course of the bachelor thesis. Moreover, the resource consumption was limited by storing the node graph externally and made individually adaptable to the user’s working memory.
The Greedy Vertex Cover algorithm’s scalability was tested on the NR database’s random samples with the newly added options. The results of DIAMOND were compared to the MMseqs2 tool, which is currently the fastest and best tool for clustering large datasets. It was shown that DIAMOND is faster than MMseqs2 when parameters are kept comparable.