News
28.06.2023
Reconstruction, validation, and phenotypic predictions strain-specific metabolic model of Staphylococcus capitis
Anna Lefarova creates a metabolic model for S. capitis in her bachelor thesis.
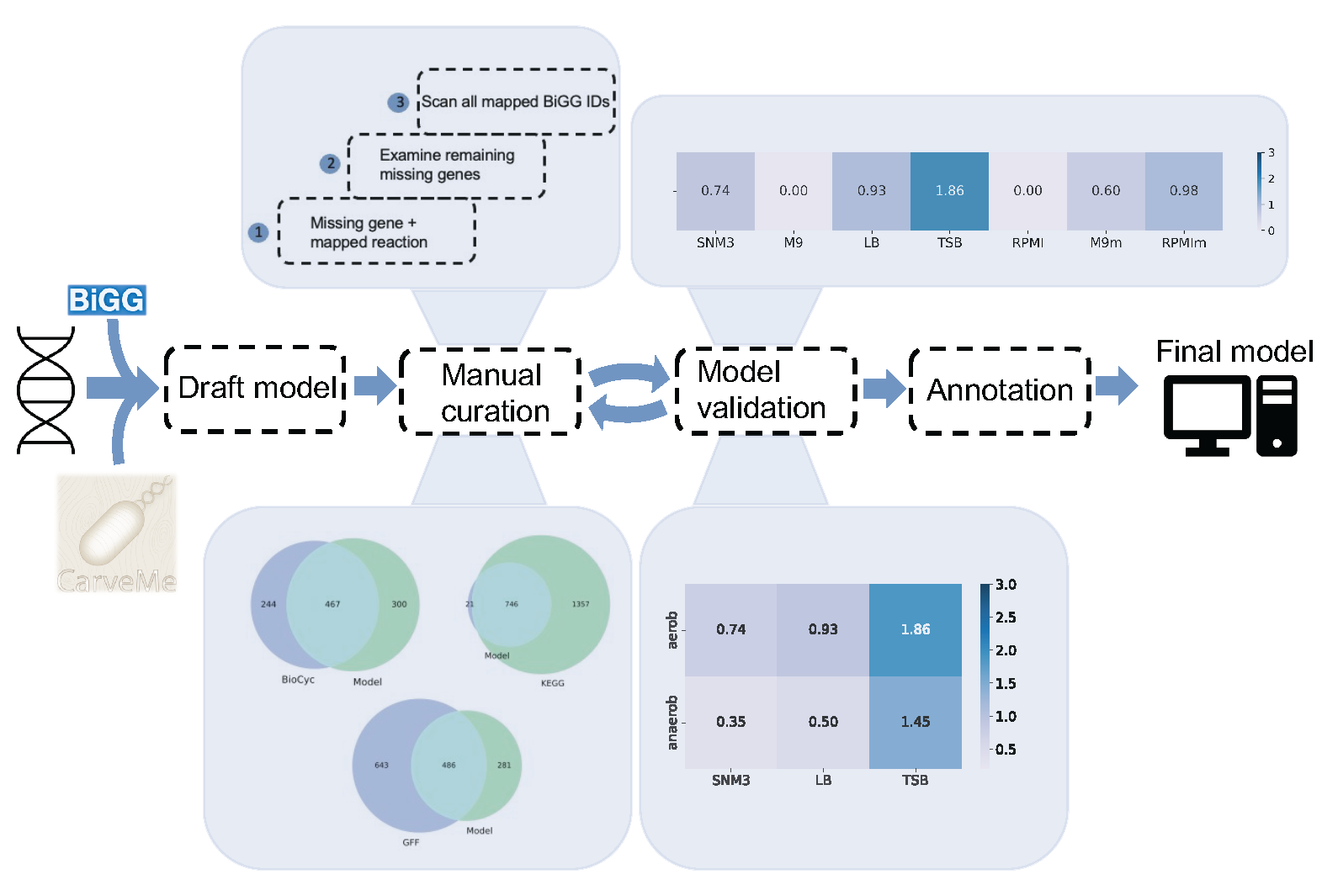
Staphylococcus capitis is a Gram-positive bacterium belonging to the coagulase-negative staphylococcus (CoNS) family. On the one hand, it is considered to have a beneficial skin microbiota due to its ability to generate an acidic pH environment and thus inhibit the growth of pathogenic microorganisms. On the other hand, clinical interest in the bacterium is increasing because it has been identified as the cause of late infections in neonatal intensive care units. The presence of S. capitis was detected on medical surfaces as well as in the nasal cavity of medical staff. The bacterium has a unique property, biofilm formation. This ability enables its survival in unfavorable environmental conditions. As part of her bachelor’s thesis, “Analysis of metabolic phenotypes of Staphylococcus capitis using restriction-based modeling,” Ms. Anna Lefarova developed a strain-specific genome-level metabolic model (GEM) of the Staphylococcus capitis organism. The computer model was built following a highly standardized protocol for genome-scale reconstructions and validated against available laboratory data. In addition, the phenotypic predictions regarding growth on the different carbon and nitrogen sources were made. The generated model was compared against automatically generated models for S. capitis. The manually curated metabolic reconstruction provides a basis for the in silico simulations. It should help to deepen the understanding of the metabolic capabilities of S. capitis.