News
26.07.2023
SPECIMEN: A workflow for automated strain-specific metabolic modeling
Carolin Brune develops a modeling toolbox and applies it to Klebsiella pneumoniae as her master thesis project.
In recent years, numerous pathogens, e.g., Klebsiella species (Klebsiella sp.), have been shown to develop resistance against antibiotics quickly. To combat these resistances, new targets and drugs for treatment need to be discovered. To assist wet lab experiments, computer models of the metabolic networks of the organism in question can be curated. However, detailed metabolic modeling to curate a high-quality model can take time due to the number of manual steps included.
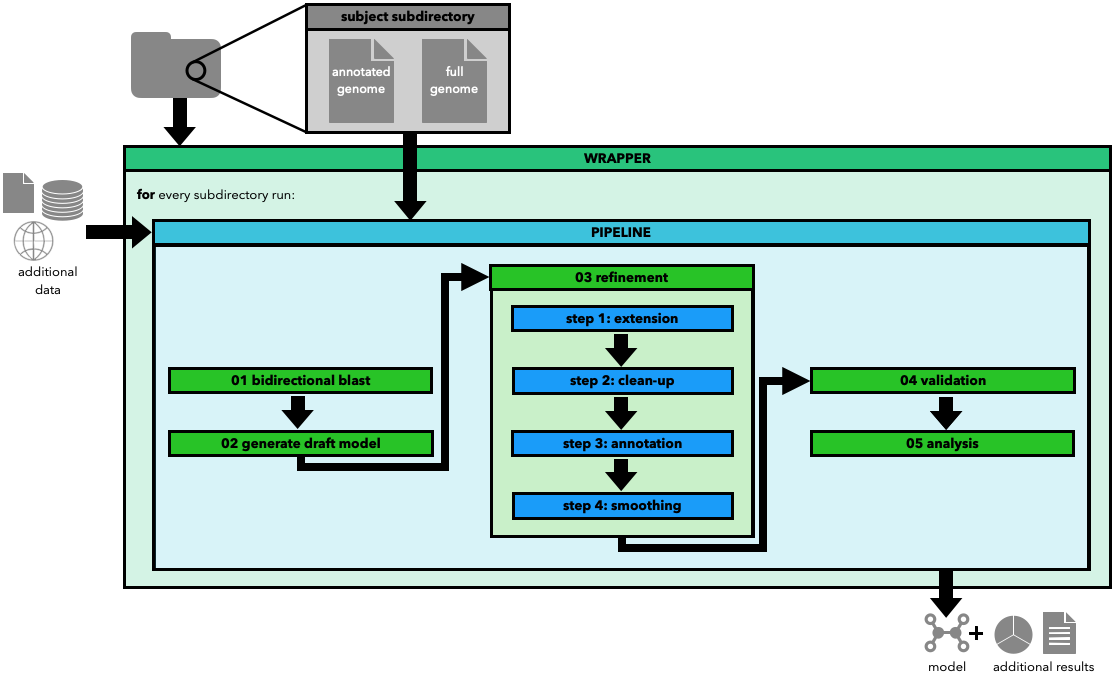
In her master thesis, Carolin Brune introduces strain-specific metabolic modeling (SPECIMEN), a Python package with an automated pipeline as its core. The workflow facilitates automated, strain-specific model curation based on a high-quality template model. Additionally, the package includes functions for further extending, refining, and analyzing the curated model, reducing the manual curation needed to create a high-quality model.
The workflow has been successfully tested using a Klebsiella pneumoniae (K. pneumoniae) genome (strain MD01) and a high-quality template model from a different strain of the same species, producing a new model for Klebsiella pneumoniae MD01.