News
17.05.2023
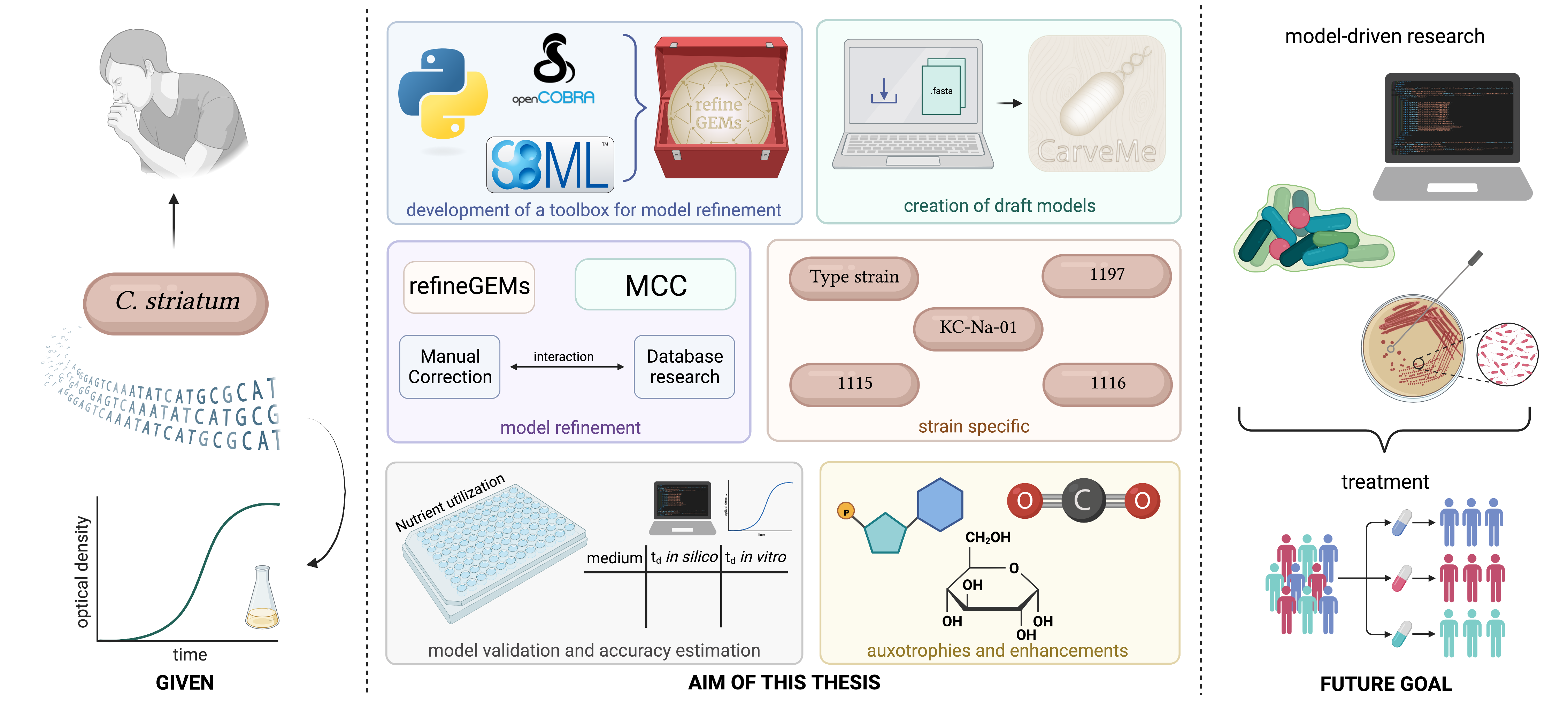
Strain-specific metabolic models of Corynebacterium striatum
Famke Bäuerle reconstructs metabolic models and validates them in vitro in the context of two master theses.
Antibiotic-resistant bacteria are leading to more and more infections worldwide. According to a Hungarian study, a disease with the bacterium Corynebacterium striatum has been associated with the need for patient ventilation even more frequently since the COVID-19 pandemic. Yet it remains largely unexplored, and the mechanisms by which it moves from being a component of the average human microbiota to a pathogen are unknown.
Metabolic models can be used to gain more insight into its metabolic behavior and potential pathogenicity factors. In a project for two master theses, Famke Bäuerle created five strain-specific genome-scale metabolic models of C. striatum and validated them using laboratory experiments. Using two experimental methods, the models provided differentially good predictions on four different media (LB, RPMI, M9, and CGXII).
In parallel, the Python package refineGEMs (github.com/draeger-lab/refinegems) was developed for other researchers to examine and curate metabolic models.
The Python package and models have already been published on BioRxiv as part of a preprint.