News
26.01.2021
Tissue-specific reconstruction of constraint-based metabolic models based on ReconX
In her master thesis, Nantia Leonidou implemented a tool for tissue-specific model reconstruction and created host-virus models relevant to the COVID-19 pandemic.
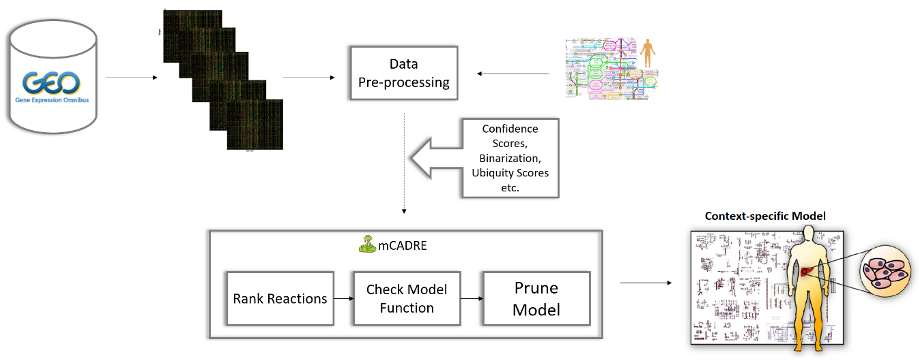
In a study published in October 2007, scientists studying coronaviruses characterized China’s situation as a ticking “time bomb” for a potential virus outbreak. They had three strong indications to worry: the animal-related eating habits in southern China, the previous appearance of SARS-CoV-like viruses in horseshoe bats, and the ability of coronaviruses to undergo recombination. Eighteen years later, the whole world experiences the realization of this prophecy with the emergence of COVID-19, one of the deadliest respiratory diseases. In this regard, scientists globally try to understand the host’s immunopathological response, how the virus adapted to new hosts, and how it spreads. Currently, great efforts are made to detect effective antiviral treatments for coronaviruses. Identifying potential antiviral targets is of great interest. One way to detect them is by analyzing metabolic changes in infected cells. In 2012, Wang et al. published mCADRE (metabolic Context-specificity Assessed by Deterministic Reaction Evaluation), aiming to reconstruct tissue-specific models using gene expression data and network topology information. The algorithm is implemented in MATLAB, and its functionality is based solely on the first version of the human model. This resulted in its limited usability in the last few years since newer and more comprehensive model versions are available.
In her master thesis, Nantia Leonidou created pymCADRE, a re-implementation of mCADRE in Python 3.8, and tested its functionality using all three currently available versions of the human metabolic network. From this, it was observed that internal optimizations done with fastFVA resulted in context-specific models closer to the ground truth.
Furthermore, viruses consume energy from the host cell to increase their mass production. Hence, they genetically re-program cells to form further virus particles and enable their reproduction. Understanding the metabolic basis of host-virus interaction could be used to predict the metabolic changes and their impacts on virus reproduction. For this reason, host-virus models (HVMs) were created with pymCADRE to help to identify potential antiviral targets against SARS-CoV-2. With those models, the existence of the recently identified potential antiviral target enzyme guanylate kinase was further verified.