Dendroscope3
An interactive viewer for rooted phylogenetic trees and networks
Researchers studying phylogenetic relationships need software that is able to visualize rooted phylogenetic trees and networks efficiently, increasingly of large datasets involving hundreds of thousands of taxa. The program should be user friendly (easy to run on all popular operating systems), facilitate interactive browsing and editing the trees and allow one to export the result in multiple file formats in publication quality. In addition, there is a need for a program that allows one to compute rooted phylogenetic networks from trees.
We have developed the platform independent tree and rooted network viewer Dendroscope that addresses these issues.
Feature List:
- Large trees with hundreds of thousands of taxa can be easily displayed, browsed and edited.
- Multiple trees and networks from a single file can be displayed together in an m by n grid
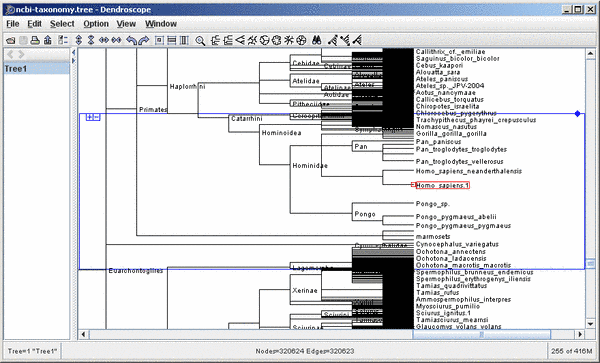
- Novel magnifying features for zooming detailed views (see screenshots)
- Find and replace tool bar that uses regular expressions
- Subtrees can be collapsed and colored
- All labels (leaves/inner nodes and edges) can be edited
- Trees can be rerooted
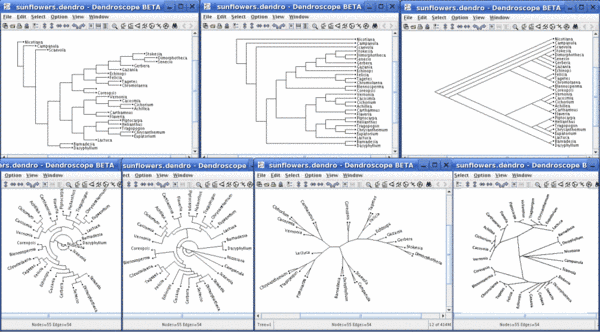
- Seven different views are available, including a rectangular, slanted, circular and radial view
- Input formats: Newick and Nexus, extended-Newick (for rooted phylogenetic networks) and Dendroscope
- Multiple graphic export formats: .eps, .svg, .png, .jpg, .gif, .bmp, .pdf
- Trees and networks can be copied and pasted between different windows
- Platform independent (Java, installers for common operating systems available)
- Consensus trees and rooted phylogenetiv networks can be computed from a set of trees
- Hybridization networks and tanglegrams for multifurcating trees on unequal taxon sets
- Commandline mode
Download
Download an installer for linux, MacOS X or Windows for version 3 Download.
(Version two is still available here).
You can test the performance of the program on the NCBI taxonomy.
References
- D.H. Huson and C. Scornavacca, Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks, Systematic Biology (2012).
- D.H. Huson, D.C Richter, C. Rausch, T. Dezulian, M. Franz and R. Rupp. Dendroscope: An interactive viewer for large phylogenetic trees, BMC Bioinformatics. 2007 Nov 22; 8(1):460 (Here you can find Table 1 from the paper in case it is wrongly displayed in the pdf or at the BMC webpage.)
The new networks capabilities introduced in version 3 are based on the following papers:
- D.H. Huson and R. Rupp, Summarizing Multiple Gene Trees using Cluster Networks, WABI 2008, 295-305.
- D.H. Huson, Drawing Rooted Phylogenetic Networks, TCBB, Jan-March 2009, Vol.6 Issue 1, pages 103-109.
- D.H. Huson, R. Rupp, V. Berry, P. Gambette and C. Paul - Computing galled networks from real data, Bioinformatics 2009 25(12):i85-i93; doi:10.1093/bioinformatics/btp217.
If you have any comments or questions then please write us an email.
Screenshots
Same tree - seven different views: Rectangular Phylogram, Rectangular Cladogram, Slanted Cladogram, Circular Phylogram, Circular Cladogram, Radial Phylogram and Radial Cladogram:
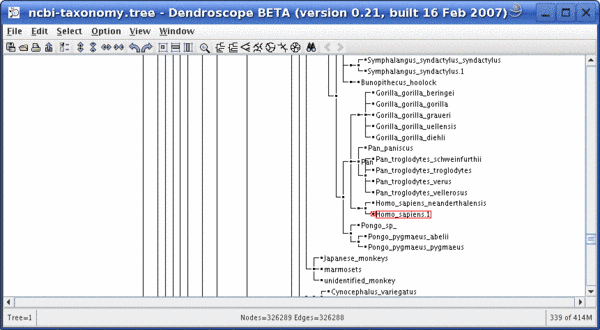
Part of the NCBI taxonomy showing Homo sapiens and his relatives without the magnifier:
Same detail of previous tree with the magnifier turned on:
Tanglegram for two trees:

